Research
Research in my lab is focussed on structural and/or biological questions that can be solved by Nuclear Magnetic Resonance. In parallel, we additionally employ a diverse range of biochemical, biomolecular and bioinformatics techniques to develop a comprehensive understanding of the studied systems.
Structural Biology
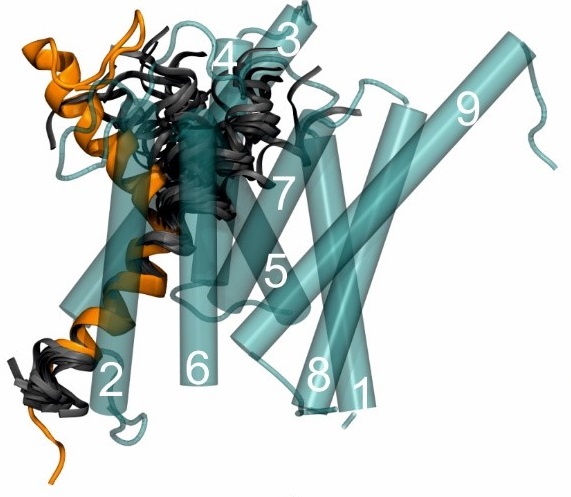
To understand cellular processes on a molecular level the 3D structures of the respective proteins have to be known. NMR spectroscopy is a versatile technique both for elaborate structure determination and for fast interaction studies e.g. for ligand screening. It is also possible to determine dynamical aspects of the studied systems at atomic resolution. We have studied a diverse range of proteins with the aim to understand how protein function is related to structure and dynamics. Current main activities of my lab center on the interplay between structure and dynamics of γ-secretase substrates, topic of the DFG funded research group FOR2290 “Understanding intramembrane proteolysis”. The intramembrane protease γ-secretase is a key player in Aβ plaque generation that occurs during Alzheimer’s disease. Studies of my lab suggest that substrate recognition is linked to the pliability of the transmembrane helices (Silber et al. 2020).
Fig 1: Docking of the structural bundle of APP-TM into a model of presenilin, the catalytic unit of γ-secretase.
Metabolomics
Metabolites are products of cellular processes that are found in organisms and body fluids. The composition of the metabolome, the sum of all metabolites, varies with many factors like the developmental state of the organism, malnutrition or disease.
We employ NMR spectroscopy paired with statistical analyses to investigate body fluids like urine or blood or extracts of cells or whole organs. In this way, we determine the chemical fingerprint of the studied systems. Our aim is to develop diagnostic tools for disease recognition or for studying the effect of chemicals or drugs on the organism. As a non-targeted approach, all sufficiently concentrated metabolites with hydrogen atoms contribute to the data.
In this way, we explored markers for acute kidney disease in children (Muhle-Goll et al. 2020) or how well blood metabolites are removed by hemodialysis (Kromke et al. 2016).
We collaborate with biologists and clinicians at KIT and the university of Heidelberg (https://www.3dmattermadetoorder.kit.edu/), who provide us with the biological and medical samples . E.g. we currently work on the identification of specific metabolites that may govern derivation of retinal organoids (https://www.3dmattermadetoorder.kit.edu/phds_771.php)
| Name | Contact |
|---|---|
| Muhle-Goll, Claudia stellvertretende Institutsleiterin, Gruppenleiterin Bio-NMR |
+49 721 608-29357 claudia muhle-goll ∂does-not-exist.kit edu CN 351 118 |
| Dopp, Franziska |
+49 721 608-29352 franziska dopp ∂does-not-exist.kit edu CN 351 114 |
| 2 weitere Personen sind nur innerhalb des KIT sichtbar. | |
Publications
Improving the Long‐term Enantioselectivity of a Silicon‐Carbon Bond‐Forming Enzyme
2025. Chemistry – A European Journal, 31 (19), Art.-Nr.: 202404688. doi:10.1002/chem.202404688
Cell-free protein production of a gamma secretase homolog
2024. Protein Expression and Purification, 215, Art.-Nr.: 106407. doi:10.1016/j.pep.2023.106407
Thio-2 Inhibits Key Signaling Pathways Required for the Development and Progression of Castration-resistant Prostate Cancer
2024. Molecular Cancer Therapeutics, 23 (6), 791–808. doi:10.1158/1535-7163.MCT-23-0354
Substrate Selection Criteria in Regulated Intramembrane Proteolysis
2024. ACS Chemical Neuroscience, 15 (7), 1321–1334. doi:10.1021/acschemneuro.4c00068
Cleavage efficiency of the intramembrane protease γ‐secretase is reduced by the palmitoylation of a substrate’s transmembrane domain
2024. The FASEB Journal, 38 (2), Art.-Nr.: e23442. doi:10.1096/fj.202302152R
Permissive Conformations of a Transmembrane Helix Allow Intramembrane Proteolysis by γ-Secretase
2023. Journal of Molecular Biology, 435 (18), 168218. doi:10.1016/j.jmb.2023.168218
Cargo Encapsulation in Photochromic Supramolecular Hydrogels Depends on Specific Guest‐Gelator Supramolecular Interactions
2023. European Journal of Organic Chemistry, 26 (20), Art.-Nr.: e202300227. doi:10.1002/ejoc.202300227
F‐Tag Induced Acyl Shift in the Photochemical Cyclization of o ‐Alkynylated N ‐Alkyl‐ N ‐acylamides to Indoles
2023. European Journal of Organic Chemistry, 26 (11), Art.-Nr.: e202201132. doi:10.1002/ejoc.202201132
Cleavage of mitochondrial homeostasis regulator PGAM5 by the intramembrane protease PARL is governed by transmembrane helix dynamics and oligomeric state
2022. Journal of Biological Chemistry, 289 (9), Art.Nr. 102321. doi:10.1016/j.jbc.2022.102321
A chemical probe for BAG1 targets androgen receptor-positive prostate cancer through oxidative stress signaling pathway
2022. iScience, 25 (5), Art.Nr. 104175. doi:10.1016/j.isci.2022.104175
Helical stability of the GnTV transmembrane domain impacts on SPPL3 dependent cleavage
2022. Scientific Reports, 12 (1), Art.-Nr.: 20987. doi:10.1038/s41598-022-24772-8
Lipase-Mediated Mechanoenzymatic Synthesis of Sugar Esters in Dissolved Unconventional and Neat Reaction Systems
2022. ACS Sustainable Chemistry & Engineering, 10 (31), 10192–10202. doi:10.1021/acssuschemeng.2c01727
Macrocyclic tetramers—structural investigation of peptide-peptoid hybrids
2021. Molecules, 26 (15), Art.Nr. 4548. doi:10.3390/molecules26154548
Lipase-Catalyzed Production of Sorbitol Laurate in a “2-in-1” Deep Eutectic System: Factors Affecting the Synthesis and Scalability
2021. Molecules, 26 (9), 2759. doi:10.3390/molecules26092759
Strain-Specific Liver Metabolite Profiles in Medaka
2021. Metabolites, 11 (11), Art.-Nr.: 744. doi:10.3390/metabo11110744
Large-Scale Recombinant Production of the SARS-CoV-2 Proteome for High-Throughput and Structural Biology Applications
2021. Frontiers in molecular biosciences, 8, Art.Nr. 653148. doi:10.3389/fmolb.2021.653148
Non-canonical Shedding of TNFα by SPPL2a Is Determined by the Conformational Flexibility of Its Transmembrane Helix
2020. iScience, 23 (12), Article: 101775. doi:10.1016/j.isci.2020.101775
The simulation of NMR data of flexible molecules: sagittamide A as an example for MD simulations with orientational constraints
2020. Physical chemistry, chemical physics, 22 (30), 17375–17384. doi:10.1039/d0cp01905d
Altered Hinge Conformations in APP Transmembrane Helix Mutants May Affect Enzyme-Substrate Interactions of γ-Secretase
2020. ACS chemical neuroscience, 11 (24), 4426–4433. doi:10.1021/acschemneuro.0c00640
Urinary NMR Profiling in Pediatric Acute Kidney Injury—A Pilot Study
2020. International journal of molecular sciences, 21 (4), Article No.1187. doi:10.3390/ijms21041187
Increased H-Bond Stability Relates to Altered ε-Cleavage Efficiency and Aβ Levels in the I45T Familial Alzheimer’s Disease Mutant of APP
2019. Scientific reports, 9 (1), 5321. doi:10.1038/s41598-019-41766-1
Trendbericht Organische Chemie: Magnetische Resonanz
2019. Nachrichten aus der Chemie, 67 (3), 71–72
Modulating Hinge Flexibility in the APP Transmembrane Domain Alters γ-Secretase Cleavage
2019. Biophysical journal, 116 (11), 2103–2120. doi:10.1016/j.bpj.2019.04.030
Efficient Extraction from Mice Feces for NMR Metabolomics Measurements with Special Emphasis on SCFAs
2019. Metabolites, 9 (3), Article: 55. doi:10.3390/metabo9030055
Trendbericht Organische Chemie 2017
2018. Nachrichten aus der Chemie, 66 (3), 249–280. doi:10.1002/nadc.20184072148
Integrated Process for the Enzymatic Production of Fatty Acid Sugar Esters Completely Based on Lignocellulosic Substrates
2018. Frontiers in Chemistry, 6, Article: 421. doi:10.3389/fchem.2018.00421
Lipase-Catalyzed Synthesis of Sugar Esters in Honey and Agave Syrup
2018. Frontiers in Chemistry, 6, Article: 24. doi:10.3389/fchem.2018.00024
Beechwood carbohydrates for enzymatic synthesis of sustainable glycolipids
2017. Bioresources and Bioprocessing, 4 (1), Art.Nr. 25. doi:10.1186/s40643-017-0155-7
Development of Bag-1L as a therapeutic target in androgen receptor-dependent prostate cancer
2017. eLife, 6, Art.Nr. e27159. doi:10.7554/eLife.27159
Organische Chemie 2016
2017. Nachrichten aus der Chemie, 65 (3), 266–304. doi:10.1002/nadc.20174059831
Sustainable enzymatic synthesis of glycolipids in a deep eutectic solvent system
2017. Journal of molecular catalysis / B, 133, 281–287. doi:10.1016/j.molcatb.2017.01.015
Untargeted multi-platform analysis of the metabolome and the non-starch polysaccharides of kiwifruit during postharvest ripening
2017. Postharvest biology and technology, 125, 65–76. doi:10.1016/j.postharvbio.2016.10.011
Organische Chemie
2016. Nachrichten aus der Chemie, 64 (3), 255–294. doi:10.1002/nadc.20164047492
Untargeted NMR Spectroscopic Analysis of the Metabolic Variety of New Apple Cultivars
2016. Metabolites, 6 (3), 29. doi:10.3390/metabo6030029
Profiling human blood serum metabolites by nuclear magnetic resonance spectroscopy: a comprehensive tool for the evaluation of hemodialysis efficiency
2016. Translational research, 171, 71–82.e9. doi:10.1016/j.trsl.2016.02.003
Influence of heating temperature, pressure and pH on recrystallization inhibition activity of antifreeze protein type III
2016. Journal of food engineering, 187, 53–61. doi:10.1016/j.jfoodeng.2016.04.019
Menthols as Chiral Auxiliaries for Asymmetric Cycloadditive Oligomerization: Syntheses and Studies of β-Proline Hexamers
2015. Organic Letters, 17 (24), 6178–6181. doi:10.1021/acs.orglett.5b03154
Glycolipids produced by Rouxiella sp. DSM 100043 and isolation of the biosurfactants via foam-fractionation
2015. AMB express, 5 (1), 82/1–11. doi:10.1186/s13568-015-0167-7
Organische Chemie 2014 (Trendbericht)
2015. Nachrichten aus der Chemie, 63, 266–305. doi:10.1002/nadc.201590092
Autoinduced catalysis and inverse equilibrium isotope effect in the frustrated Lewis pair catalyzed hydrogenation of imines
2015. Chemistry - a European journal, 21, 8056–8059. doi:10.1002/chem.201500805
Extracellular aromatic biosurfactant produced by Tsukamurella pseudospumae and T. spumae during growth on n-hexadecane
2015. Journal of Biotechnology, 211, 107–114. doi:10.1016/j.jbiotec.2015.06.424
Lipase-catalyzed synthesis of glucose-6-O-hexanoate in deep eutectic solvents
2015. European journal of lipid science and technology, 117 (2), 161–166. doi:10.1002/ejlt.201400459
Structure of the Membrane Anchor of Pestivirus Glycoprotein Erns, a Long Tilted Amphipathic Helix
2014. PLoS pathogens, 10 (2), Art. Nr.: e1003973. doi:10.1371/journal.ppat.1003973
Trehalose lipid biosurfactants produced by the actinomycetes Tsukamurella spumae and T. pseudospumae
2014. Applied microbiology and biotechnology, 98 (21), 8905–8915. doi:10.1007/s00253-014-5972-4
Coregulator Control of Androgen Receptor Action by a Novel Nuclear Receptor-Binding Motif
2014. Journal of biological chemistry, 289 (13), 8839–8851. doi:10.1074/jbc.M113.534859
Organische Chemie 2013. Magnetische Resonanz
2014. Nachrichten aus der Chemie, 62, 282–283
Dendrimer-type peptoid-decorated hexaphenylxylenes and tetraphenylmethanes: Synthesis and structure in solution and in the gas phase
2014. Chemistry - a European journal, 20, 16273–16278. doi:10.1002/chem.201404024
Influence of Freezing and Storage Procedure on Human Urine Samples in NMR-Based Metabolomics
2013. Metabolites, 3 (2), 243–258. doi:10.3390/metabo3020243
Alternating asymmetric self-induction in functionalized pyrrolidine oligomers
2013. Angewandte Chemie - International Edition, 52 (48), 12736–12740. doi:10.1002/anie.201302862
Organische Chemie 2012
2013. Nachrichten aus der Chemie, 61 (3), 265–297. doi:10.1002/nadc.201390087
Trendberichte Organische Chemie: Magnetische Resonanz
2013. Nachrichten aus der Chemie, 61, 295
Structural Characterization of a Peptoid with Lysine-like Side Chains and Biological Activity using NMR and Computational Methods
2013. Organic and Biomolecular Chemistry, 11 (4), 640–647. doi:10.1039/c2ob27039k
A Peptidic Unconjugated GRP78/BiP Ligand Modulates the Unfolded Protein Response and Induces Prostate Cancer Cell Death
2012. PLoS one, 7 (10), Art.Nr. e45690. doi:10.1371/journal.pone.0045690
Structural investigation of the transmembrane segment of the PDGF receptor beta and the oncoprotein E5 by circular dichoroism and NMR
2012. Biophysical Society 56th Annual Meeting, San Diego, Calif., February 25-29, 2012 Biophysical Journal, 102(2012) No.3 Suppl.1, a (Abstract)
Structural investigations of the heterotetrameric E5/PDGF-receptor β complex by oriented circular dichroism and solid state NMR
2012. Biophysical Society 56th Annual Meeting, San Diego, Calif., February 25-29, 2012
Rapid calculation of protein chemical shifts using bond polarization theory and its application to protein structure refinement
2012. Physical chemistry chemical physics, 14 (35), 12263–12276. doi:10.1039/c2cp41726j
Hydrophobic matching controls the tilt and stability of the dimeric platelet-derived growth factor receptor (PDGFR) β transmembrane segment
2012. The journal of biological chemistry, 287 (31), 26178–2686. doi:10.1074/jbc.M111.325555
Structure analysis of the membrane-bound PhoD signal peptide of the Tat translocase shows an N-terminal amphiphilic helix
2012. Biochimica et biophysica acta / Biomembranes, 1818 (12), 3025–3031. doi:10.1016/j.bbamem.2012.08.002
Bag-1 proteins inhibit prostate cancer cell growth through a distinct sequence that binds the glucose regulated protein GRP78
2011. 3rd Internat.Meeting on Cell Stress and Apoptosis - to Arturo, Salerno, I, June 23-25, 2011
Pro-apoptotic properties of Bag-1 peptides in the control of prostate cancer cell growth
2011. 102nd Annual Meeting of the American Association for Cancer Research (AACR), Orlando, Fla., April 2-6, 2011 Cancer Research, 71(2011) Nr.8, Suppl.1 Abstract 5403
Structure analysis of the prePhoD signal peptide: functional implications of an amphiphilic helix for approaching the Tat translocase
2010. 24th Internat.Conf.on NMR in Biological Systems, Cairns, AUS, August 22-27, 2010
Structure of the pore-forming subunit of the twin arginine translocase determined by combining liquid- and solid-state NMR
2010. 24th Internat.Conf.on NMR in Biological Systems, Cairns, AUS, August 22-27, 2010
The Glc₂Man₂-fragment of the N-glycan precursor - a novel ligand for the glycan-binding protein malectin?
2010. Organic and Biomolecular Chemistry, 8, 3294–99. doi:10.1039/C004502K
Structural role of the conserved cysteines in the dimerization of the viral transmembrane oncoprotein E5
2010. Biophysical Journal, 99, 1764–72. doi:10.1016/j.bpj.2010.06.073
Short cationic antimicrobial peptides interact with ATP
2010. Antimicrobial Agents and Chemotherapy, 54, 4480–83. doi:10.1128/AAC.01664-09
Structure of the pore-forming subunit of the twin arginine translocase determined by combining liquid- and solid-state NMR
2010. World Wide Magnetic Resonance 2010 : Joint EUROMAR 2010 and 17th ISMAR Conf., Firenze, I, July 4-9, 2010
The strictly conserved Cys76 plays a crucial role in the conformational stability of reduced Px III
2010. EMBO Conf.’Catalytic Mechanisms by Biological Systems : at the Interface Between Chemistry and Biology’, Hamburg, May 5-7, 2010
Analysis of the specific interactions between the lectin domain of malectin and diglucosides
2010. Glycobiology, 20, 1010–20. doi:10.1093/glycob/cwq059
Structural features of the transmembrane E5 oncoprotein from bovine papillomavirus
2010. Biophysics of Membrane-Active Peptides : 455th WE-Heraeus-Seminar, Bad Honnef, April 11-14, 2010
Structural characterization of the transmembrane helix of the platelet-derived growth factor receptor-β
2010. Biophysics of Membrane-Active Peptides : 455th WE-Heraeus-Seminar, Bad Honnef, April 11-14, 2010
The conserved Cys76 plays a crucial role for the conformation of reduced glutathione peroxidase-type tryparedoxin peroxidase
2010. FEBS Letters, 584, 1027–32. doi:10.1016/j.febslet.2010.01.054
Malectin - a novel lecitin of the endoplasmic reticulum and a candidate new player in the early steps of protein N-glycosylation
2009. Annual Meeting of the Society for Glycobiology, Fort Worth, Tex., November 12-15, 2008 Glycobiology, 18(2008) (Abstract)
Malectin - a novel lecitin of the endoplasmic reticulum and a candidate new player in the early steps of protein N-glycosylation
2009. Annual Meeting of the Society for Glycobiology, Fort Worth, Tex., November 12-15, 2008 Glycobiology,(2008) (Abstract), 18, 948
Interactions of the Tat-dependent signal peptide of B.subtilis with the Tat-translocase
2009. CFN Summer School on Nano-Biology, Bad Herrenalb, September 8-11, 2009
Influence of the conserved cysteines on the membrane structure of the bovine papillomavirus E5 oncoprotein
2009. Signal Transduction and Disease : Trinat.Fall Meeting 2009 of the Societies of Biochemistry and Molecular Biology of Belgium, Germany and the Netherlands, Aachen, September 27-30, 2009
Role of the transmembrane helix of plateled-derived growth factor β receptor
2009. Signal Transduction and Disease : Trinat.Fall Meeting 2009 of the Societies of Biochemistry and Molecular Biology of Belgium, Germany and the Netherlands, Aachen, September 27-30, 2009
Structure and dynamics of the human muscle LIM protein
2009. FEBS Letters, 583, 1017–22. doi:10.1016/j.febslet.2009.02.021
Malectin - a novel carbohydrate-binding protein of the endoplasmic reticulum and a candidate player in the early steps of protein N-glycosylation
2009. 3rd EU-NMR Annual User Meeting, Autrans, F, January 26-29, 2009
Structural characterization of the Tat-translocase pore forming protein TatAsub(d) from B. subtilis
2008. 8th Internat.Conf.on Magnetic Resonance in Biological Systems, San Diego, Calif., August 24-29, 2008
¹H, ¹³C, and ¹⁵N assignment of the oxidized and reduced forms of T.brucei glutathione peroxidase-type tryparedoxin peroxidase
2008. Biomolecular NMR Assignments, 2, 65–68. doi:10.1007/s12104-008-9086-8
Competition saturation transfer difference experiments improved with isotope editing and filtering schemes in NMR-based screening
2008. Journal of the American Chemical Society, 130, 17148–153. doi:10.1021/ja804468k
Structural basis for a distinct catalytic mechanism in trypanosoma brucei tryparedoxin peroxidase
2008. Journal of Biological Chemistry, 283, 30401–11. doi:10.1074/jbc.M803563200
On the significance of Toc-GTPase homodimers
2008. Journal of Biological Chemistry, 283, 23104–12. doi:10.1074/jbc.M710576200
Malectin: A novel carbohydrate-binding protein of the endoplasmic reticulum and a candidate player in the early steps of protein n-glycosylation
2008. Molecular Biology of the Cell, 19, 3404–14. doi:10.1091/mbc.E08-04-0354
Non empirical computation of protein NMR properties - a rapid and accurate approach using bond polarization theory
2008. 8th Triennial Congress of the World Association of Theoretical and Computational Chemists (WATOC 2008), Sydney, AUS, September 14-19, 2008
Study of the catalytic cycle of GPX3 from trypanosoma brucei
2008. EU-NMR Annual User Meeting, Königstein, January 23-25, 2008

 We have studied a diverse range of proteins with the aim to understand how protein function is related to structure and dynamics. Current main activities of my lab center on the interplay between structure and dynamics of γ-secretase substrates, topic of the DFG funded research group
We have studied a diverse range of proteins with the aim to understand how protein function is related to structure and dynamics. Current main activities of my lab center on the interplay between structure and dynamics of γ-secretase substrates, topic of the DFG funded research group