Metabolomics
Metabolites are products of cellular processes that are found in organisms and body fluids. The sum of all metabolites are called the metabolome. Its composition varies with many factors like the developmental state of the organism, malnutrition or disease.
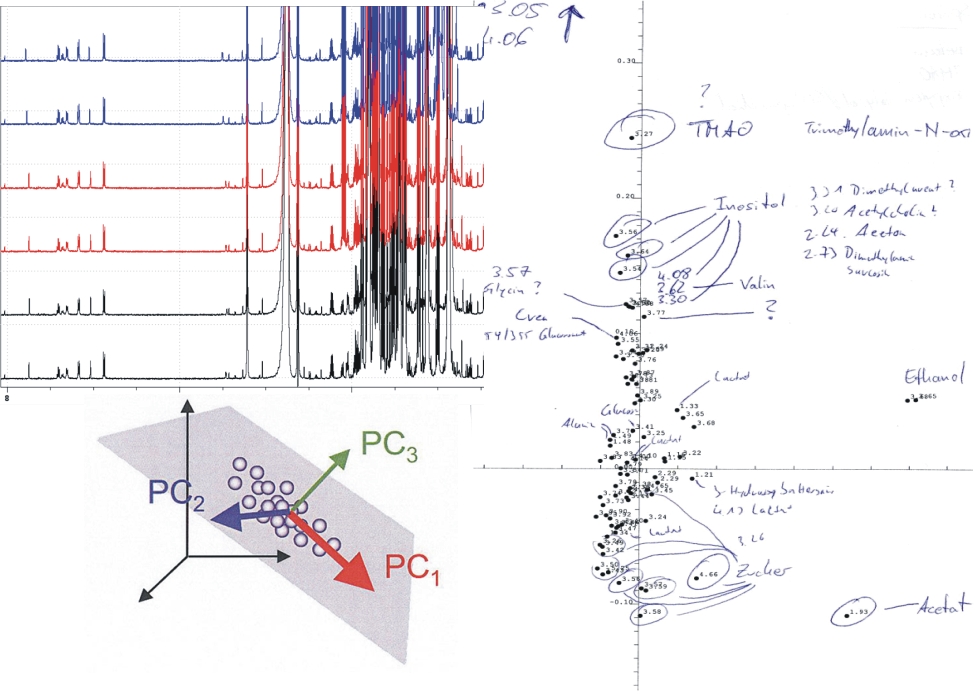
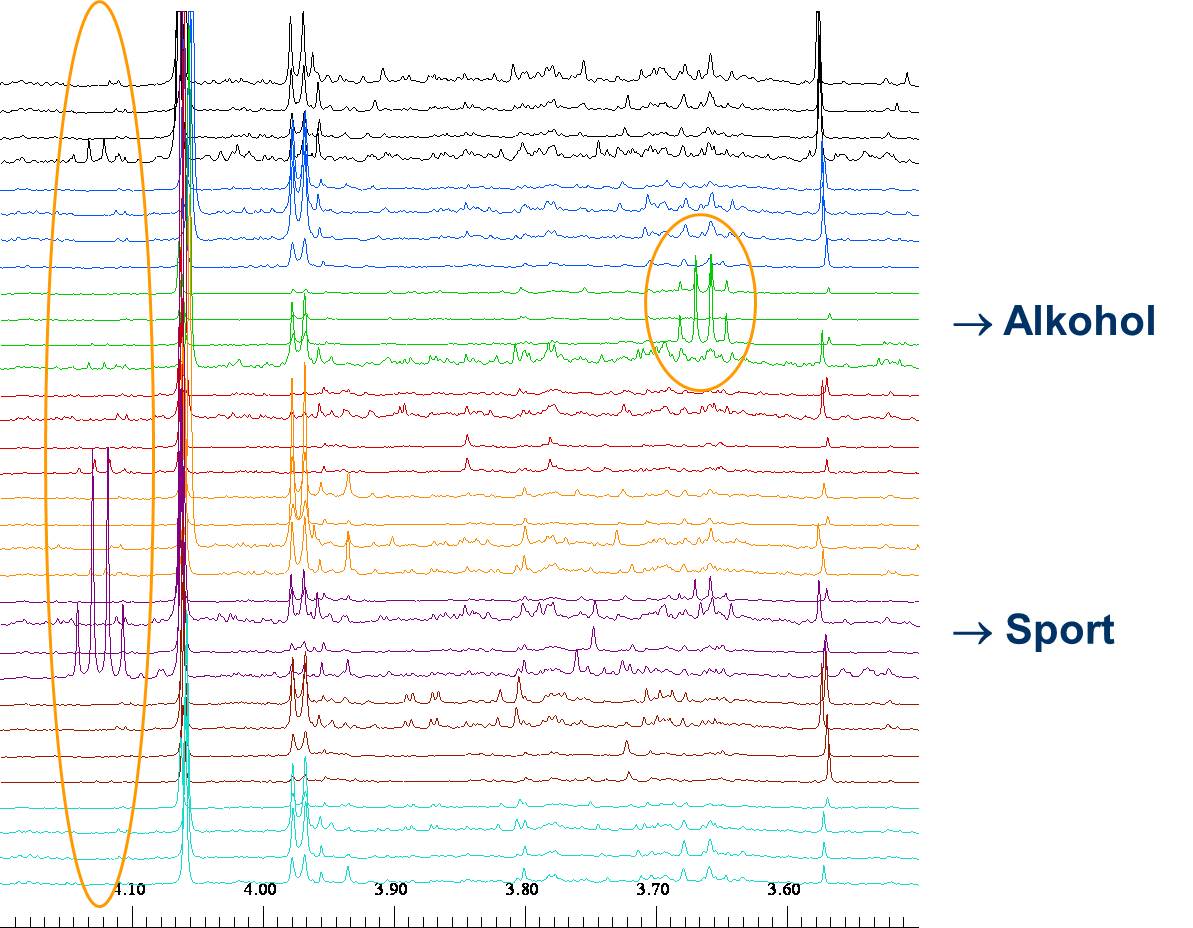
Metabolomics deals with the systematic investigation of body fluids like urine, blood or cell liquids or even whole organs and organisms. The metabolome is regarded as a chemical fingerprint of the organism that can be exploited as a diagnostic tool for disease recognition or for studying the effect of chemicals or drugs on the organism. NMR spectroscopy is one of the main techniques employed in metabolomics studies, because of the manifold of signals that can be detected over a large dynamic range of several orders of magnitude and the inherent quantification of metabolites by spectral integration. Hundreds of components and their variation between different samples can be analysed in a few minutes/hours with high accuracy in a non-destructive way. As a non-targeted approach, all sufficiently concentrated metabolites with hydrogen atoms contribute to the spectrum. NMR therefore can be used as a non-invasive technique in vitro and in vivo that provides metabolic information otherwise lost upon extraction. Univariate und multivariate statistical (e.g. principal component analysis) analyses are mandatory to manage the large data volume generated during experiments with the representative high number of samples.
Zebrafish
Zebrafish (Danio rerio) are widely used model organisms because they are vertebrates and many scientific observations can be transferred to higher vertebrates like humans. The embryos of the zebrafish develop outside of the mother animal which makes it easy to manipulate them with microinjections of transplantations. Additionally, a huge set of mutants is available and their circadian clocks can be easily manipulated with a light, dark cycle.
In collaboration with Thomas Dickmeis of the ITG, KIT we investigate the effect of missing cortisol levels on the metabolism of the zebrafish over the day. Therefore, we measure extracts of wild-type and mutant zebrafish larvae that were collected at different times during one day. After identifying the differences, we try to normalize the metabolic profiles of the phenotype by treating the zebrafish with drugs.
In collaboration with Nicholas Foulkes of the ITG, KIT we measure the rhythmicity of metabolites in zebrafish livers. Later, this approach should be transferred to cavefish that show very different behavior, depending on the habitat they live in.
Dialysis
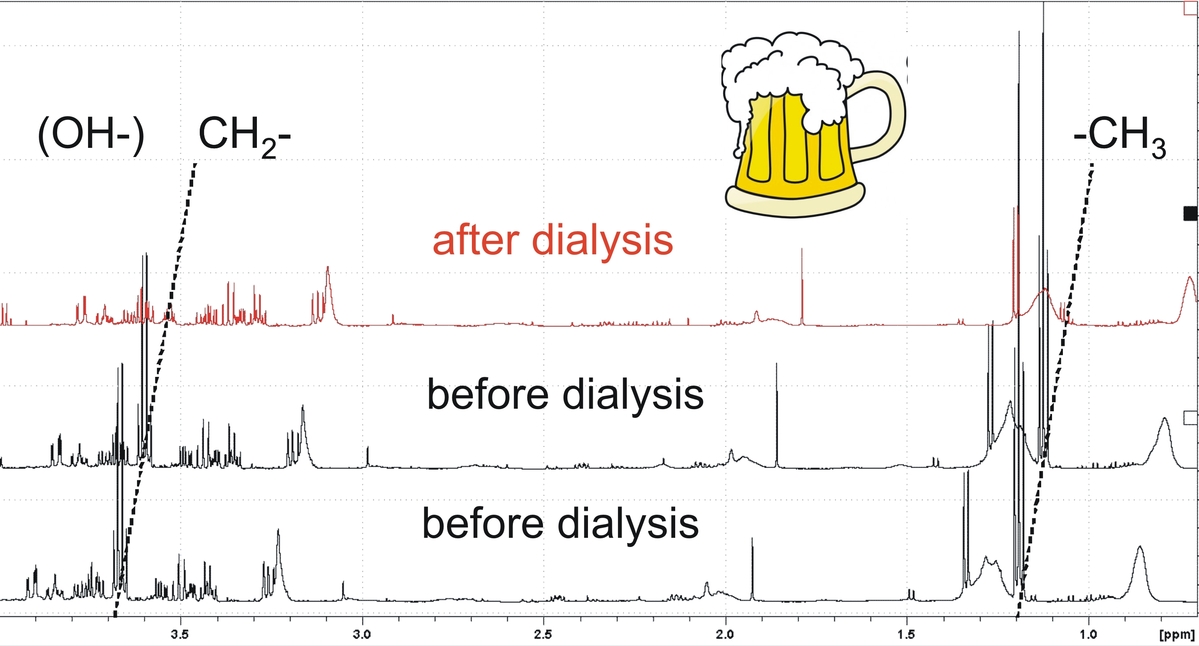
Kidney insufficiency is a multi-cause disease that is treated by blood dialysis. In between the bi- or triweekly hemodialysis sessions metabolites accumulate in the blood plasma. Efficiency of hemodialysis is monitored by measuring the renal clearance for which urea levels before and after treatment are compared. Creatinin or glucose blood concentrations that are also monitored are blood metabolites that can be routinely detected in clinical lab practice by a number of diagnosis methods. However, experience shows that the level of these small metabolites is not sufficient to predict longterm outcome of treatment.
In collaboration with the Städtische Klinikum Karlsruhe we have explored how variable the accumulation of metabolites in the blood of a cohort of chronic kidney disease (CKD) patients is and how well the metabolites are removed by hemodialysis. We have compared the blood of CKD patients taken prior and immediately after hemodialysis over a time period of four months. Both the concentration of metabolites and the efficiency of dialysis vary between the patients but over the period of four months the individual behaviour is comparably constant.
Plants
Food metabolomics investigates food extracts. Metabolomics can be used as tool to differentiate between plant species or to investigate the influence of different cultivation sites. It can also be used as quality control by generating unique fingerprints of the different species. Identification of specific marker metabolites or compound patterns within aqueous or sometimes hydrophobic extracts of plant material helps to understand the underlying differences. Identified metabolites can be correlated with observations like disease resistances, phenotype, fruit flavor or ripening to provide insight into plant metabolism and even in plant taxonomy.
Metabolomics has been applied to plants used in traditional Chinese medicine in collaboration with the Nick group at the Institute of Botany, KIT. Chinese medicine is not well explored with respect to active ingredients and sometimes is potentially dangerous due to similarly looking toxic plants, which can be mistaken for the medicinal plants. We have used metabolomics to analyse crude drug preparations for their content in medicinal plants, and used this information for identification of the otherwise sometimes almost indistinguishable plants extracts. In fruit metabolomics, we have studied tomato, kiwi and apples in collaboration with the Max-Rubner Institute Karlsruhe.
HuMet
The Human Metabolome (HuMet) study was performed at the Technische Universtität München (TUM). The aim was to investigate the normal range of metabolite concentrations and the variability in the reaction of humans to nutritional and other challenges. Therefore, over the time course of two times two days, 15 well characterised healthy young men underwent 6 challenges, including 36 h of fasting, an oral glucose tolerance test, an oral lipid tolerance test and a physical activity test. Blood and urine samples were collected frequently and analysed by NMR, LC-MS and other analytical methods. The study showed that for some of the determined metabolites variability increased after challenges, whereas for others it decreased, which corresponded with the anabolic and catabolic state of the volunteers, respectively. Further analysis of the data is in progress.
KarMeN
KarMeN is the acronym for “Karlsruhe Metabolomics and Nutrition” which is a cross-sectional human metabolomics study investigating the determinants of the plasma and urine metabolome of healthy adults and was performed at the Max Rubner-Institut (Federal Research Institute of Nutrition and Food) in Karlsruhe. More than 300 men and women in the age range of 18 – 80 years were recruited, anthropometric, functional and clinical parameters were determined, and dietary intake was assessed. Blood and urine samples were collected and analysed by several targeted and untargeted metabolomics techniques, including NMR. Using appropriate bioinformatics and biostatistics methods, the aim will be to find metabolites or metabolite patterns that are associated with age, sex, physical fitness or diet of the study participants.
Contact M. Rist.